mikeb and I were discussing the papers given at BioConcur the
other day, and we were puzzling out the differences between the
standard way of modelling biochemical and biological reactions
(including genetic interference reactions) and our “alternative”
modelling style. I suppose the “alternative” style must have been
tried before, and that it must display some obvious weakness we
haven’t spotted yet.
Here’s a comparison of the two styles:
| Biological Systems Element |
Standard Model |
"Alternative" Model |
| Molecule | Process | Message |
| Molecule species | — | Channel |
| Interaction Capability | Channel | Process |
| Interaction | Communication | Movement (and computation) |
| Modification |
State and/or channel change |
State and/or channel change |
| Reaction Rate |
attached to channel |
attached to prefix |
Join and Rates
We’ve been thinking about using a stochastic π-variant with
a join operator. Having join available allows the expression
of state machines to be much more direct than in a joinless stochastic
π. We started modelling the lambda-switch to fit the model from this
BioConcur paper, and saw that it seems more natural to attach the
reaction rate to each individual join rather than to each channel as
BioSPI does.
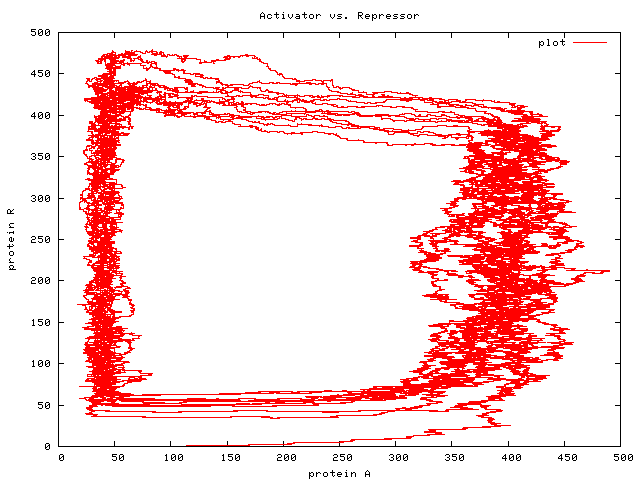
Bisimulation
The standard model has comm meaning interaction; we have comm
meaning movement in some sense: atoms (mass) move from one
locus (molecular species) to another, or a molecule undergoes
conformational change, which is a kind of motion from one state or
class or species to another. This means that bisimulation in the
alternative model has to do with movement rather than interaction
— which seems more fine-grained, but perhaps less natural.
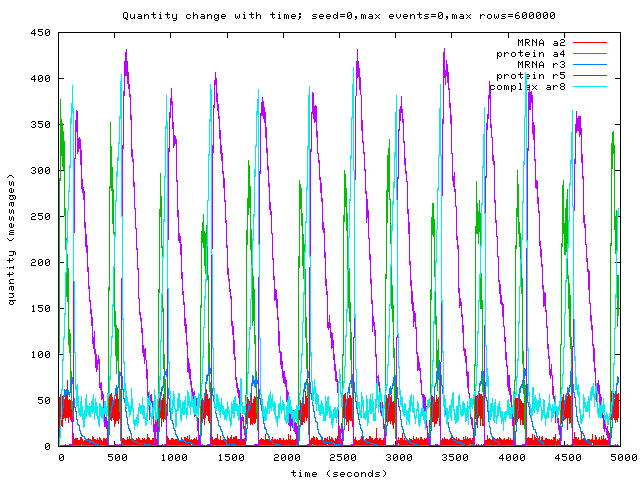
Uncertainty in Biological Simulation
We also thought about a few properties biological simulation
systems ought to have. One important one is a notion
of uncertainty. Few of the current systems are honest about
their limits — only one of the papers at BioConcur displayed
error bars on any of its graphs! The systems in use all generate
probable trajectories through state-space, and this is I assume
implicitly to be understood by the reader of the various program
outputs, but it would be useful to have the uncertainty in the results
reflected in the various displays that these systems produce.
Besides the uncertainties that come from stochastic modelling and
Monte-Carlo-type approaches in general, all the inputs to the systems
(eg. rates) are purely empirical data, with associated uncertainties,
and these uncertainties are not propagated through the system. Not
only do the outputs not show the variation in results due to random
sampling, but the outputs do not explore the limits of the accuracy of
the inputs, either.
Reagent concentration vs. Gillespie vs. Modelling reagent state
The alternative model deals well with high reagent concentrations,
since a high concentration is modelled as a large number of messages
outstanding on a channel. The Gillespie scheduling algorithms are
O(n)-time in the number of enabled reactions, not in the
concentrations of the reagents. The standard model suffers here
because each molecule is modelled as a process — that is, as an
enabled reaction. In a naïve implementation, this makes the scheduling
complexity O(n)-time (or at best, O(log(n)) time if the
Gibson-Bruck algorithm is used) in the number of molecules in the
system.
However, modelling molecules as processes seems to provide one
advantage that doesn’t easily carry over into the alternative model:
molecule-processes can involve internal state that any interacting
molecule need not be concerned with. For example, enzyme-private
channels with different rates associated with them might be used to
model the different reactivities of phosphorylated and
unphosphorylated states of the enzyme.
Perhaps a hybrid of the two models will be workable; after all,
even in the alternative model, you have the freedom of the restriction
operator available for implementing complex behaviours. The
alternative model allows some common patterns to be expressed more
simply than the standard joinless model though, it seems.
We’re still thinking about ways of keeping the
molecules-as-messages approach while allowing reactions to specialise
on molecule-message state or to ignore it, as appropriate.
Compartments
BioSPI 3.0 took the standard stochastic-π implementation and added
BioAmbients to it (see the 2003 paper by Regev, Panina, Silverman,
Cardelli and Shapiro). Any stochastic-join will probably benefit from
being able to model compartments similarly.
The simple, ambientless approach suffers some of the same problems
as SBML is facing with regard to recognising the similarity of species
across compartments. Some way of factoring out reactions/processes/etc
common across all compartments should be useful.